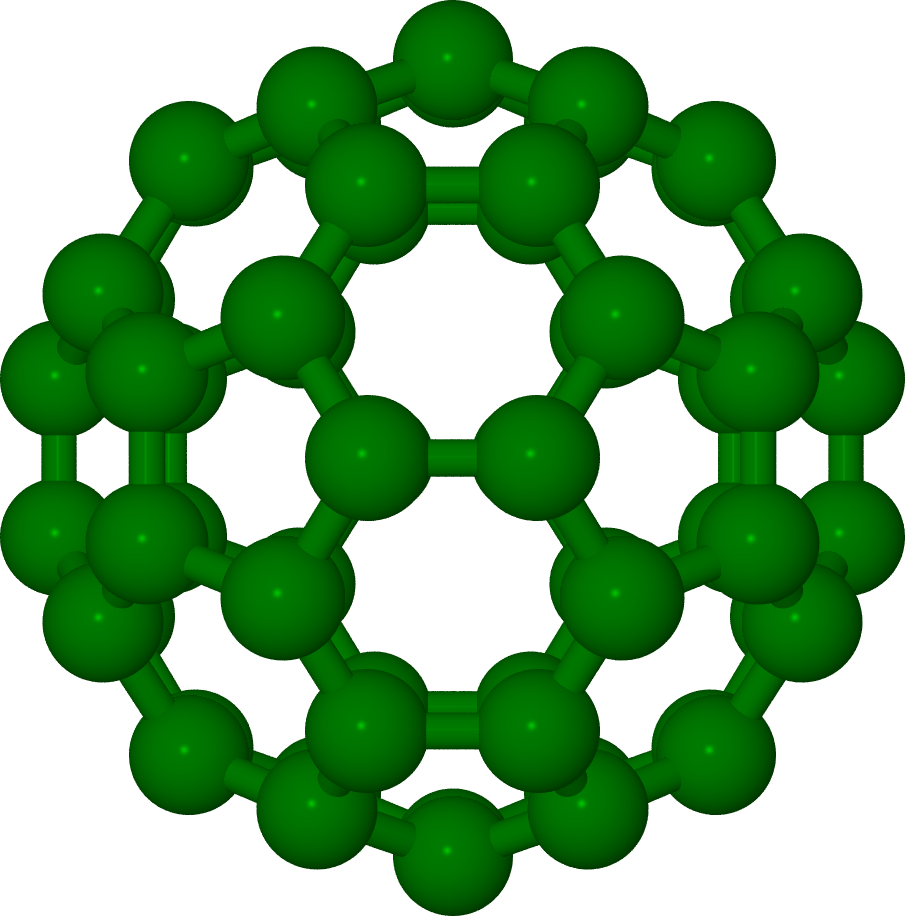
Tutorial 3 Electronic structure of the C60 molecule¶
This tutorial will explain use of NRLMOL to study the electronic structure of C60 molecule. The C60 molecule is spherical carbon molecule - also called bucky ball. It was found in the laboratory at Rice university in 1985 by Harold Kroto, James R. Heath, Sean O’Brien, Robert Curl, and Richard Smalley. The 1996 nobel prize was awarded to Kroto, Curl and Smalley for the discovery of C60 and related molecules. This molecule posses very high symmetry- icosahedral symmetry.
We will make use of this symmetry in our calculations. All carbon atoms in C60 are equivalent to each other, that is, once the position of one carbon atom is specified the position of remaining carbon atoms can be obtained using the point group operations of the icosahedral group. This tutorial will explain how to accomplish this and make use of symmetry to simplify the calculations.
We assume that the code us compiled with appropriate set of parameters needed for arrays. The following set of parameters will work.
IMPLICIT REAL*8 (A-H,O-Z)
PARAMETER ( MX_PROC= 40)
PARAMETER ( MXSPN= 1)
PARAMETER ( MAX_PTS= 100000)
PARAMETER ( MAX_FUSET= 2)
PARAMETER ( MAX_IDENT= 10)
PARAMETER ( MX_CNT= 60)
PARAMETER ( MXATMS= MX_CNT )
PARAMETER ( NMUPMAX= 7)
PARAMETER ( MAX_BARE= 30)
PARAMETER ( MAX_CON= 12)
PARAMETER ( MAX_OCC= 2400)
PARAMETER ( MAXUNSYM= 55)
PARAMETER ( NDH= 1500)
PARAMETER ( NDH_TOT= 1126500)
PARAMETER ( MX_GRP= 120)
PARAMETER ( MAX_REP= 10)
PARAMETER ( MAXSLC= 3300)
PARAMETER ( LOCMAX= 360)
PARAMETER ( ISMAX= 32)
PARAMETER ( MAXSYMSALC= 32)
PARAMETER ( MXRPSP= 100)
PARAMETER ( MXPTAB= 100)
PARAMETER ( MXLPSP= 2)
PARAMETER ( MXKPT= 1)
PARAMETER ( MPBLOCK= 100)
PARAMETER ( MXPOISS= 100)
PARAMETER ( NSPEED= 25)
PARAMETER ( LDIM= 3)
PARAMETER ( CUTEXP= 40.00000000000000 )
PARAMETER ( MX_SPH= 1000)
PARAMETER ( MAX_VIRT_PER_SYM= 500)
PARAMETER ( MAXGFIT= 15)
PARAMETER ( MAXLSQF= 50)
PARAMETER ( MTEMP= 60000 )
PARAMETER ( KRHOG= 10 )
As you know by now, CLUSTER file is the main input file of NRLMOL. For C60 the CLUSTER file is
GGA-PBE*GGA-PBE (DF TYPE EXCHANGE*CORRELATION)
IH (TD, OH, IH, X, Y, XY, ... OR GRP)
1 (NUMBER OF INEQUIV. ATOMS IN CH4)
0.000000 6.579993 1.321695 6 ALL
0.0 0.0 (NET CHARGE AND NET SPIN)
Note that we have only one atom listed and the point group is listed to be Ih. If you are using point group symmetry then only positions of inequivalent atoms need to be listed. The NRLMOL will generate full geometry of the system using the point group operations. It is strongly recommended that you visualize the geometry generated - XMOL.DAT or XMOL.xyz files using molecular viewer such as JMOL or MOLEKEL or any other of your choice.
- Now run the calculation.
$ rm -f GEOCNVRG SYMBOL
$ ./cluster > print.SCF
Open the GEOCNVRG file and write down the energy. It should be
1.0000000000000000E-003
CONVERGE TRUE
ENERGY= -2284.358327861622
TOTAL GRADIENT= 6.9943783981219765E-005
LARGEST NUCLEAR GRADIENT= 6.9943783981219765E-005
and the forces are
-2284.3583278616 1 Energy and Symbol NCALC
0.00000000 6.57999300 1.32169500 6 6
0.000000000000D+00 -0.934955152640D-05 -0.693160789707D-04 <--- MAX-FORCE
-0.213162820728D-13 0.710542735760D-14 0.213162820728D-13
0.000000000000D+00 0.000000000000D+00 0.000000000000D+00
Note that forces on only one atom (inequivalent) are listed.
The eigenvalues are stored in file called “EVALUES”. Please inspect it. Look at the degeneracy of the eigenvalues. Identify HOMO, LUMO etc. The file is also displayed below for your convenience.
This is an abridged file containing eigenvalues of C60.
********* NEW TRY ************, SPIN: 1
REP: 1 DIM: 1 NUMBER OF BASES: 25
1 25
-9.942805 -0.8997024 -0.5549701 -0.4479115 -0.1991965E-01
0.6689239E-01 0.1684883 0.4065947 0.4303353 0.4932246
0.6197493 0.8237995 1.064748 1.332207 1.589216
.
.
.
REP: 2 DIM: 3 NUMBER OF BASES: 60
3 60
-9.942771 -9.942283 -0.8811058 -0.6511179 -0.4751674
-0.4263302 -0.1540616 0.2963368E-01 0.7381854E-01 0.1399013
0.1454381 0.2194449 0.2953231 0.4100078 0.4159666
0.5064494 0.5198368 0.5662745 0.6077245 0.6927512
0.8485903 0.9020563 0.9212636 0.9778088 1.120618
.
.
.
REP: 3 DIM: 5 NUMBER OF BASES: 95
5 95
-9.942707 -9.942522 -9.942192 -0.8455306 -0.7306929
-0.5751889 -0.4086972 -0.3857767 -0.3299505 -0.2612101
-0.7485659E-01 0.5984700E-01 0.9606631E-01 0.1289818 0.1606308
0.2017960 0.2727632 0.3200607 0.3361940 0.3987439
0.4505640 0.4721895 0.4830797 0.4905669 0.5863982
0.5938349 0.6509424 0.6840220 0.6948194 0.8087404
0.8537736 0.9215714 0.9419371 0.9592563 0.9766250
.
.
.
REP: 4 DIM: 3 NUMBER OF BASES: 60
3 60
-9.942610 -9.942173 -0.8087070 -0.6119900 -0.4521482
-0.3383461 -0.8228363E-01 0.8985531E-01 0.1057484 0.1731967
0.2351114 0.3167932 0.4029469 0.4170402 0.4388469
0.5078198 0.5737927 0.6229951 0.7266742 0.7936461
0.9151430 0.9266100 0.9995002 1.073048 1.199648
.
.
.
REP: 5 DIM: 4 NUMBER OF BASES: 70
4 70
-9.942629 -9.941986 -0.7771414 -0.4280769 -0.3666027
-0.3181925 0.1970728E-01 0.8832295E-01 0.1412645 0.2453531
0.3710384 0.3930982 0.4018634 0.4195257 0.5115655
0.5622322 0.5934607 0.6458433 0.7175404 0.7465492
0.7555818 0.8274507 0.9549796 1.034820 1.078526
.
REP: 6 DIM: 4 NUMBER OF BASES: 70
4 70
-9.942545 -9.942070 -0.7135614 -0.5127091 -0.3953816
-0.2569762 -0.5496157E-02 0.1097230 0.1575782 0.2465148
0.3105353 0.4047911 0.4485227 0.4531293 0.4763553
0.4850277 0.5600762 0.6488439 0.6656025 0.7337152
0.8110428 0.9402047 0.9808577 1.030000 1.105739
REP: 7 DIM: 5 NUMBER OF BASES: 80
5 80
-9.942488 -9.942127 -0.6595450 -0.4869222 -0.3186715
-0.2144125 -0.2052547E-01 0.1355438 0.2234478 0.2350119
0.3204446 0.3928607 0.3957610 0.4817556 0.4930063
0.5036710 0.5642048 0.5714468 0.7307460 0.7358558
0.7466057 0.8818037 0.9426629 0.9976558 1.047811
1.088609 1.136037 1.226505 1.297072 1.401576
2.099996 2.127763 2.167014 2.206178 2.279180
REP: 8 DIM: 3 NUMBER OF BASES: 45
3 45
-9.942249 -0.5708627 -0.1140607 0.1309249 0.1794275
0.3661029 0.4497591 0.4917586 0.5693141 0.6491583
0.8200522 0.9194278 0.9646669 1.101807 1.220654
1.351788 1.480114 1.581128 1.767750 1.927398
1.952246 2.024065 2.094322 2.217394 2.285956
REP: 9 DIM: 3 NUMBER OF BASES: 45
3 45
-9.941977 -0.3861205 0.3803237E-01 0.2679375 0.3120214
0.4249118 0.4853449 0.5541875 0.6313738 0.6528077
0.8270208 0.8480218 1.054382 1.105491 1.275696
1.421811 1.585232 1.855841 1.955594 1.993680
2.065630 2.158184 2.230132 2.310101 2.441308
REP: 10 DIM: 1 NUMBER OF BASES: 10
1 10
0.6175634 0.9026151 1.484611 2.284678 2.714255
3.303272 3.605378 5.801911 5.971903 6.756511
FERMI LEVEL: -0.1842370492849899 TEMP: 1.0000000000000000E-004
SUMMARY OF EVALUES AND THEIR OCCUPANCIES:
1 REP: 1 DEG: 1 SPIN: 1 ENERGY: -9.94281 OCC: 1.00000
2 REP: 2 DEG: 3 SPIN: 1 ENERGY: -9.94277 OCC: 1.00000
3 REP: 3 DEG: 5 SPIN: 1 ENERGY: -9.94271 OCC: 1.00000
4 REP: 5 DEG: 4 SPIN: 1 ENERGY: -9.94263 OCC: 1.00000
5 REP: 4 DEG: 3 SPIN: 1 ENERGY: -9.94261 OCC: 1.00000
6 REP: 6 DEG: 4 SPIN: 1 ENERGY: -9.94254 OCC: 1.00000
7 REP: 3 DEG: 5 SPIN: 1 ENERGY: -9.94252 OCC: 1.00000
8 REP: 7 DEG: 5 SPIN: 1 ENERGY: -9.94249 OCC: 1.00000
9 REP: 2 DEG: 3 SPIN: 1 ENERGY: -9.94228 OCC: 1.00000
10 REP: 8 DEG: 3 SPIN: 1 ENERGY: -9.94225 OCC: 1.00000
11 REP: 3 DEG: 5 SPIN: 1 ENERGY: -9.94219 OCC: 1.00000
12 REP: 4 DEG: 3 SPIN: 1 ENERGY: -9.94217 OCC: 1.00000
13 REP: 7 DEG: 5 SPIN: 1 ENERGY: -9.94213 OCC: 1.00000
14 REP: 6 DEG: 4 SPIN: 1 ENERGY: -9.94207 OCC: 1.00000
15 REP: 5 DEG: 4 SPIN: 1 ENERGY: -9.94199 OCC: 1.00000
16 REP: 9 DEG: 3 SPIN: 1 ENERGY: -9.94198 OCC: 1.00000
17 REP: 1 DEG: 1 SPIN: 1 ENERGY: -0.899702 OCC: 1.00000
18 REP: 2 DEG: 3 SPIN: 1 ENERGY: -0.881106 OCC: 1.00000
19 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.845531 OCC: 1.00000
20 REP: 4 DEG: 3 SPIN: 1 ENERGY: -0.808707 OCC: 1.00000
21 REP: 5 DEG: 4 SPIN: 1 ENERGY: -0.777141 OCC: 1.00000
22 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.730693 OCC: 1.00000
23 REP: 6 DEG: 4 SPIN: 1 ENERGY: -0.713561 OCC: 1.00000
24 REP: 7 DEG: 5 SPIN: 1 ENERGY: -0.659545 OCC: 1.00000
25 REP: 2 DEG: 3 SPIN: 1 ENERGY: -0.651118 OCC: 1.00000
26 REP: 4 DEG: 3 SPIN: 1 ENERGY: -0.611990 OCC: 1.00000
27 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.575189 OCC: 1.00000
28 REP: 8 DEG: 3 SPIN: 1 ENERGY: -0.570863 OCC: 1.00000
29 REP: 1 DEG: 1 SPIN: 1 ENERGY: -0.554970 OCC: 1.00000
30 REP: 6 DEG: 4 SPIN: 1 ENERGY: -0.512709 OCC: 1.00000
31 REP: 7 DEG: 5 SPIN: 1 ENERGY: -0.486922 OCC: 1.00000
32 REP: 2 DEG: 3 SPIN: 1 ENERGY: -0.475167 OCC: 1.00000
33 REP: 4 DEG: 3 SPIN: 1 ENERGY: -0.452148 OCC: 1.00000
34 REP: 1 DEG: 1 SPIN: 1 ENERGY: -0.447911 OCC: 1.00000
35 REP: 5 DEG: 4 SPIN: 1 ENERGY: -0.428077 OCC: 1.00000
36 REP: 2 DEG: 3 SPIN: 1 ENERGY: -0.426330 OCC: 1.00000
37 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.408697 OCC: 1.00000
38 REP: 6 DEG: 4 SPIN: 1 ENERGY: -0.395382 OCC: 1.00000
39 REP: 9 DEG: 3 SPIN: 1 ENERGY: -0.386121 OCC: 1.00000
40 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.385777 OCC: 1.00000
41 REP: 5 DEG: 4 SPIN: 1 ENERGY: -0.366603 OCC: 1.00000
42 REP: 4 DEG: 3 SPIN: 1 ENERGY: -0.338346 OCC: 1.00000
43 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.329950 OCC: 1.00000
44 REP: 7 DEG: 5 SPIN: 1 ENERGY: -0.318671 OCC: 1.00000
45 REP: 5 DEG: 4 SPIN: 1 ENERGY: -0.318192 OCC: 1.00000
46 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.261210 OCC: 1.00000
47 REP: 6 DEG: 4 SPIN: 1 ENERGY: -0.256976 OCC: 1.00000
48 REP: 7 DEG: 5 SPIN: 1 ENERGY: -0.214412 OCC: 1.00000
49 REP: 2 DEG: 3 SPIN: 1 ENERGY: -0.154062 OCC: 0.000000E+00
50 REP: 8 DEG: 3 SPIN: 1 ENERGY: -0.114061 OCC: 0.000000E+00
51 REP: 4 DEG: 3 SPIN: 1 ENERGY: -0.822836E-01 OCC: 0.000000E+00
52 REP: 3 DEG: 5 SPIN: 1 ENERGY: -0.748566E-01 OCC: 0.000000E+00
To be added: Discussion of orbital degeneracies- plot of wave functions